Vector DB performance issues are like debugging a distributed system designed by someone who hates you. Everything affects everything else, and the error messages tell you absolutely nothing useful.
HNSW: Fast But Hungry
HNSW promises fast searches but will eat your RAM for breakfast. I think I'm running like... a million-ish 1536-dimensional vectors? Maybe 1.2 million? Anyway, it's using something like 6.2GB of memory - most of that is just storing the vectors, but the index overhead adds up fast. When you run out of RAM, performance doesn't just slow down, it falls off a fucking cliff because everything starts swapping to disk.
Pinecone's docs explain the theory if you want to dig deeper (though their examples assume you have infinite memory). Brendan Gregg wrote about why CPU metrics lie during memory pressure - turned out that's exactly what was happening to us.
I learned this the hard way during a product demo where queries went from like 8-10ms to... Jesus, I think it was around 25-30 seconds? Something ridiculously slow. Turns out we hit the memory limit and Linux started swapping. The demo was a complete disaster and I looked like a fucking idiot in front of the whole engineering team.
The Linux kernel docs have good info on memory management concepts, and Redis has solid guidance on vector performance optimization.
IVF: Clustering Hell
IVF indexes are supposed to be smart - they cluster your vectors and only search the relevant clusters. The theory says use √N clusters, but real data doesn't give a shit about theory.
Text embeddings cluster in completely fucked up ways. I had this dataset where like... shit, I think 70-something percent of vectors ended up in just 3 or 4 clusters? Maybe it was 75%? Hard to remember exactly, but it was bad. They were all similar content so of course they clustered together. Queries to those clusters were slow as hell while empty clusters did nothing. Ended up using way more clusters than the theory said we needed.
OpenAI has some blog posts about their embedding improvements (though they never mention the clustering problems). There's research on cluster imbalance but it's pretty academic.
Product Quantization: Lossy But Necessary
PQ compresses vectors massively - like 90-95% compression ratios. A 1536-dimensional vector that takes 6KB becomes something like... I don't know, 150-200 bytes? Maybe less. Sounds great until you realize the accuracy hit might not be worth it.
Facebook Engineering has some FAISS compression posts (the math is a nightmare). FAISS docs cover the basics if you want to suffer through them.
Spent like 3 days, maybe 4, optimizing PQ parameters only to find out our similarity thresholds were completely fucked for the compressed data. Users were getting garbage results and we had to roll back the "optimization." That was a fun conversation with the product team.
GPU Lies and Marketing Bullshit
Everyone told me GPU acceleration would make everything faster. Spoiler alert: it doesn't, at least not for single queries. The overhead of moving data to the GPU eats up any speed gains unless you're processing hundreds of queries at once.
Single queries on our A100 were actually slower than CPU because of the PCIe transfer time. Only batch queries over... I think it was like 64 or 128? Either way, only big batches showed real improvements. And batch processing doesn't help when users want real-time responses.
NVIDIA's marketing is all "GPU acceleration!" but they don't mention the PCIe tax. Their CUDA docs explain the memory bottlenecks if you can wade through the marketing bullshit.
Memory vs Disk: The 1000x Problem
When your index fits in RAM, queries are fast. When it doesn't, they're slow as fuck. There's no middle ground.
RAM access takes nanoseconds, NVMe SSDs take microseconds - that's a 1000x difference that shows up directly in your query times. Memory-based searches finish in a few milliseconds, disk-based ones can take hundreds of milliseconds.
The Dimensionality Curse
Here's something nobody tells you: high-dimensional vectors are weird. Once you get above 1000 dimensions, all vectors start looking equally similar. The curse of dimensionality isn't just theory - it actually breaks similarity search.
I've seen embeddings where the "closest" match was barely closer than a random vector. OpenAI's 1536-dimensional embeddings are total overkill for most use cases. Reducing to 768 or even 384 dimensions often gives better results with way less computation. Wish I'd known that before we deployed the 1536 version to prod.
Distance Metric Performance Gotchas
Cosine similarity without pre-normalized vectors is a performance killer. If you're computing the magnitude every time, you're doing 3x more work than necessary.
Pre-normalize your vectors during ingestion and cosine similarity becomes as cheap as dot product. This optimization was huge for us - like 40-50% faster queries? Maybe more? Definitely felt stupid for not doing it from the start.
Real Performance Killers
Connection pools: When your pool runs out, queries start queuing. Latency goes to shit even though your CPU and memory look fine. Monitor active connections, not just system resources.
Memory fragmentation: HNSW indexes get slower over time as memory gets fragmented. We had to restart our service every week just to defragment memory and get performance back.
Bad query patterns: Random access patterns destroy cache performance. If your queries are all over the place, you're guaranteed to have cache misses and slow searches.
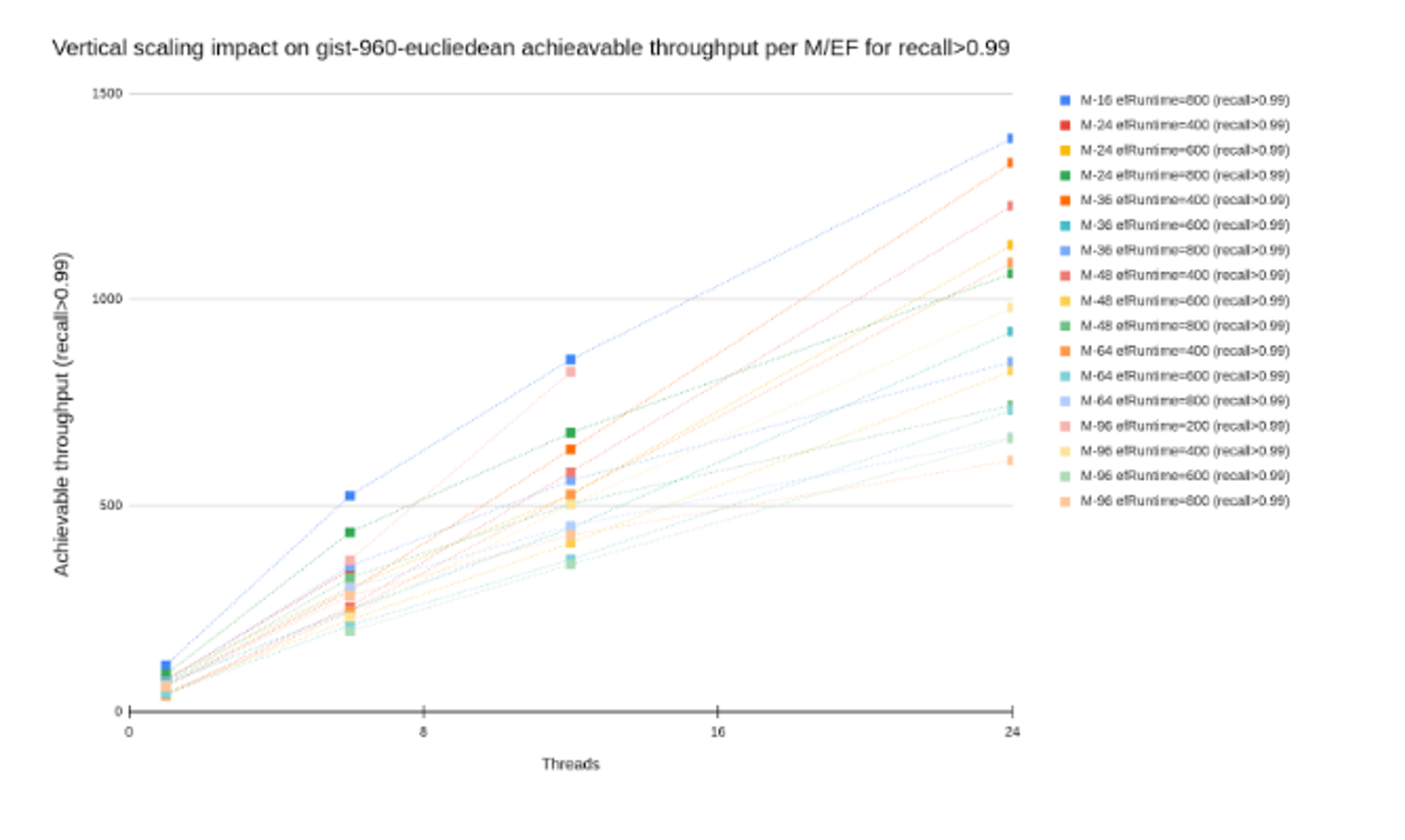
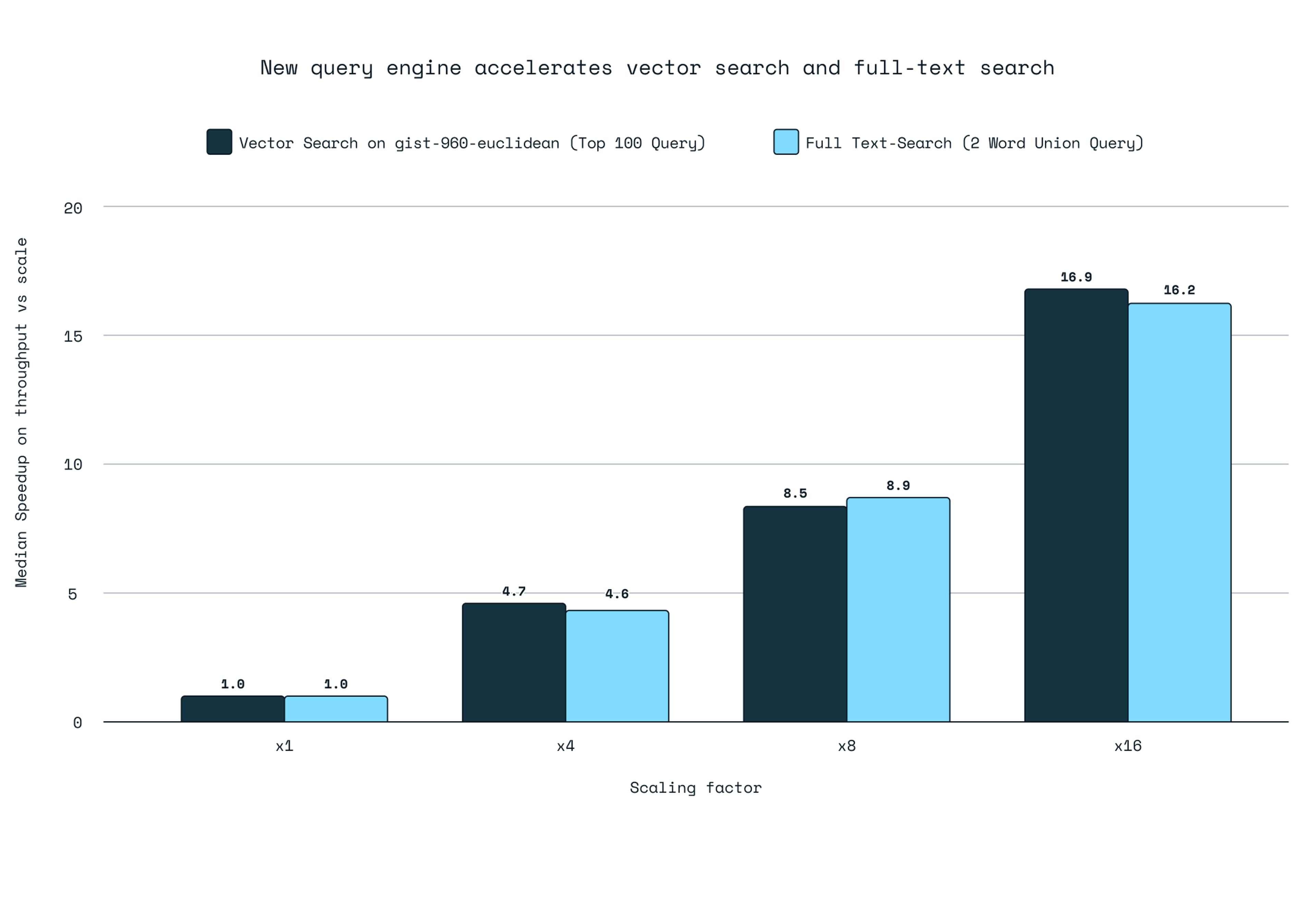
Concurrent queries: Most vector DBs suck at concurrent searches. They'll serialize on some internal data structure and your multi-core machine performs like a single-core potato.
The real lesson? Every optimization comes with tradeoffs, and marketing benchmarks are complete bullshit. Test everything with your actual data or you'll get burned.