I've watched pandas kill more production systems than any other Python library. The pattern is always the same: works perfectly in development, explodes spectacularly in production when data volume doubles.
Why pandas Eats Memory Like Candy
pandas wasn't designed for big data. It loads your entire dataset into RAM, then makes copies for every operation. That innocent-looking df.groupby() can triple your memory usage instantly.
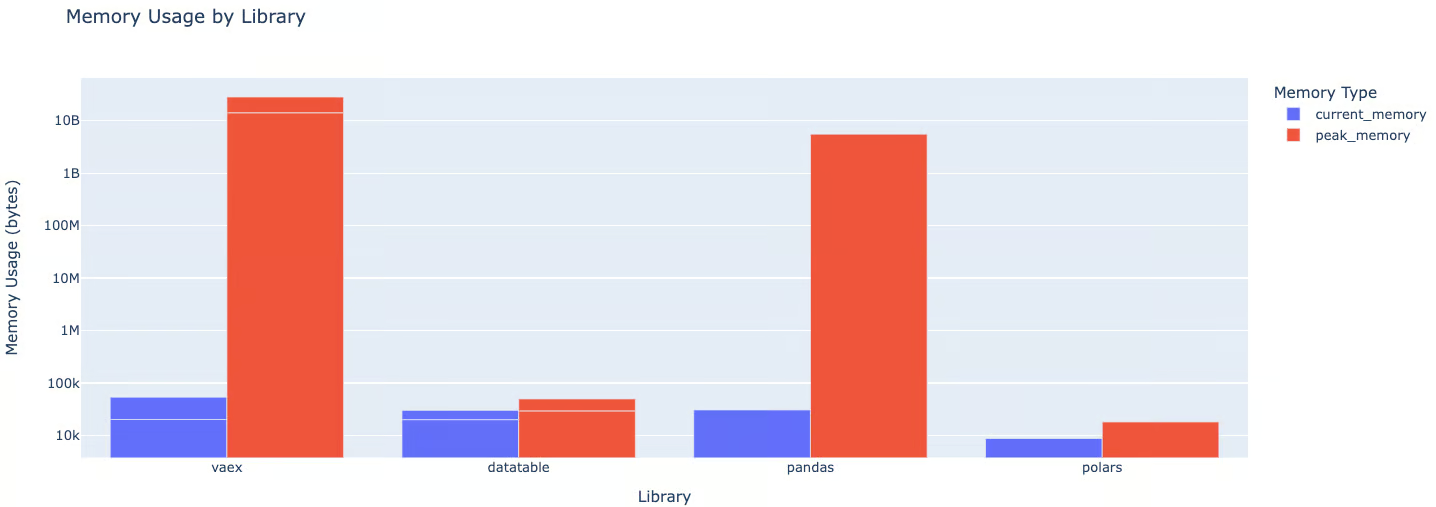
pandas uses 1100x more memory than Polars and 29x more than DataTable - this is why your containers keep getting OOMKilled
Here's what actually happens when you load a 2GB CSV:
- Initial load: 2GB file → 6GB DataFrame (text-to-numeric conversion overhead)
- Type inference: Another 2GB copy while pandas figures out column types
- First operation: Yet another copy, now you're at 12GB+ RAM usage
The worst part? pandas operations aren't atomic. If you run out of memory halfway through a join, you've lost everything and need to start over.
The Production Reality Check
Netflix: They handle this by chunking everything. Their ETL pipelines never process more than 100MB at once in pandas.
JPMorgan: They use specialized data type optimization and convert everything to categories/numeric codes before processing.
Airbnb: They switched critical paths to Spark/PySpark for anything over 1GB.
The pattern is clear: successful pandas deployments at scale require aggressive memory management and fallback strategies. Check the pandas memory optimization guide and performance enhancement documentation for official recommendations.
Memory Optimization That Actually Works
1. Data Type Optimization (30-80% memory reduction)
## This function saved my ass multiple times
def optimize_dtypes(df):
for col in df.select_dtypes(include=['int64']).columns:
if df[col].min() > -128 and df[col].max() < 127:
df[col] = df[col].astype('int8')
elif df[col].min() > -32768 and df[col].max() < 32767:
df[col] = df[col].astype('int16')
for col in df.select_dtypes(include=['float64']).columns:
df[col] = df[col].astype('float32')
return df
2. Categorical Data (50-90% reduction for repeated strings)
If you have a column with repeated values (like country codes), convert it to categorical:
df['country'] = df['country'].astype('category')
I once reduced a 12GB DataFrame to 2GB just by making string columns categorical. The performance improvement was ridiculous.
3. Chunked Processing (Infinite scale, finite patience)
When all else fails, process your data in chunks:
chunk_size = 10000
results = []
for chunk in pd.read_csv('massive_file.csv', chunksize=chunk_size):
processed_chunk = chunk.groupby('category').sum()
results.append(processed_chunk)
final_result = pd.concat(results).groupby(level=0).sum()
This pattern has saved my career at least twice.
For more advanced optimization techniques, check out these resources:
- pandas memory profiling with memory_profiler
- Modin for parallel pandas operations
- Dask for distributed computing
- Polars as a fast alternative
- Vaex for out-of-core processing
The PyData Stack Exchange and pandas GitHub issues are goldmines for real-world performance solutions.